We use and provide training for classical virological assays as well as state of the art digital PCR analysis, nanopore sequencing, infectious (BSL-3) live cell imaging, and high throughput screening assays.
A549-AT infection model for monitoring SARS-CoV-2 replication
Using the Sleeping Beauty transposon system (Kowarz et al. 2015), we established a reliable and adaptable cellular infection model based on A549 lung cells. This model facilitated our high-throughput SARS-CoV-2 infection studies and enabled live cell imaging at a significantly reduced cost.
Read the paper to find out more: Widera et al. 2021 Front Microbiol. 2021 Jul 29:12:701198. doi: 10.3389/fmicb.2021.701198. eCollection 2021.
You might also read our interview on Generating a cellular infection model for monitoring SARS-CoV-2 replication in the Tecan Journal: external link
Nanopore based sequencing of viral genomes
Oxford Nanopore Technology (ONT) is revolutionizing our lab by enabling real-time, long-read sequencing of viral genomes without requiring expensive equipment, making advanced genomic analysis more accessible. Its portable devices can sequence directly from complex samples, streamlining workflows and facilitating high-resolution viral surveillance in both clinical and environmental samples.

(Foto: M. Widera)
Our use of ONT ranges from plasmid verification to assessing the genomic integrity of our in vitro-produced virus stocks, as well as characterizing viral sequences in wastewater.
Quantitative digital PCR
We use digital PCR to detect low-abundance viral nucleic acids in wastewater and to accurately quantify viral genome copy equivalents. This method delivers unparalleled sensitivity by partitioning samples into thousands of individual reaction compartments, allowing rare DNA or RNA molecules to be detected and quantified with high precision. Additionally, digital PCR eliminates the need for standard curves, offering absolute quantification that enhances reproducibility and streamlines experimental workflows.
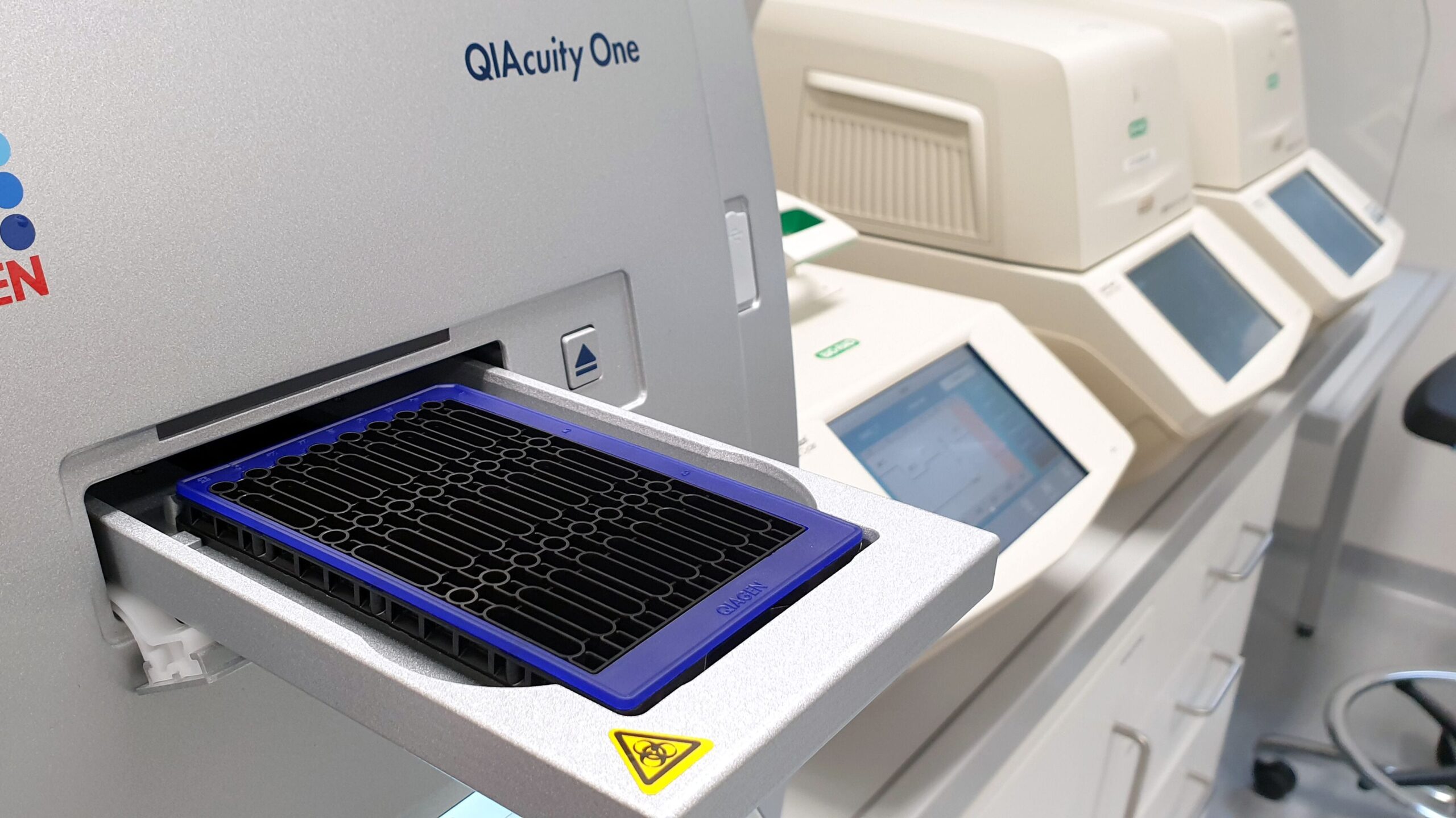
(Foto: M. Widera)
Generation of stable reporter cell lines
Under construction…
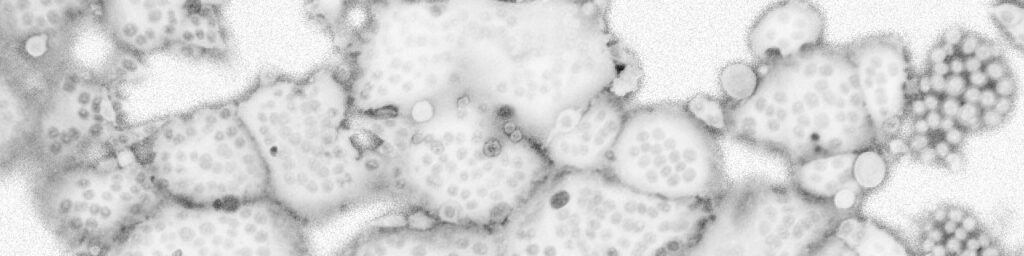
(Foto: M. Widera)
Infectious cell culture models and live cell imaging
Under construction…
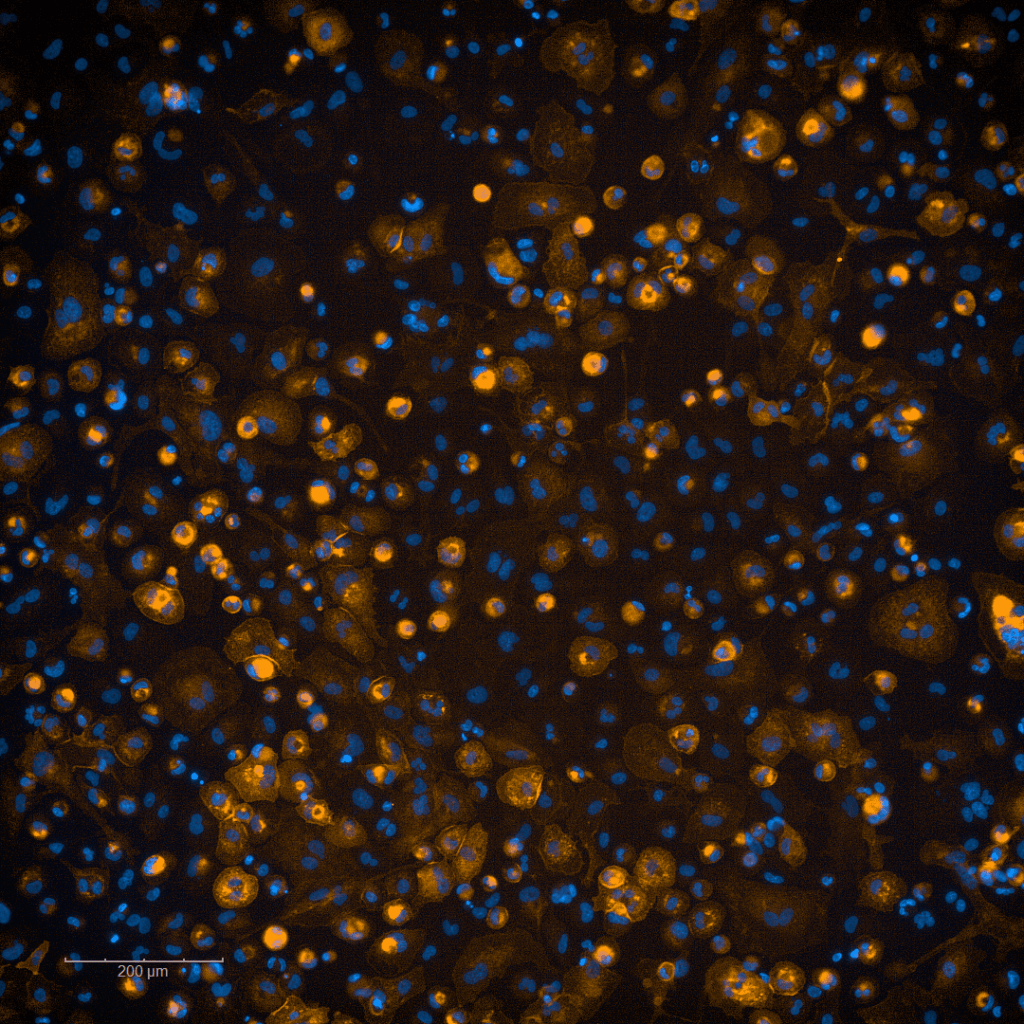
(Foto: F. Roesmann)
Isolation of viral nucleic acids from wastewater samples
Under construction…
(Foto: M. Widera)